Question: How are open reading frames or exons predicted from genomic sequence? A. If the open reading fram…
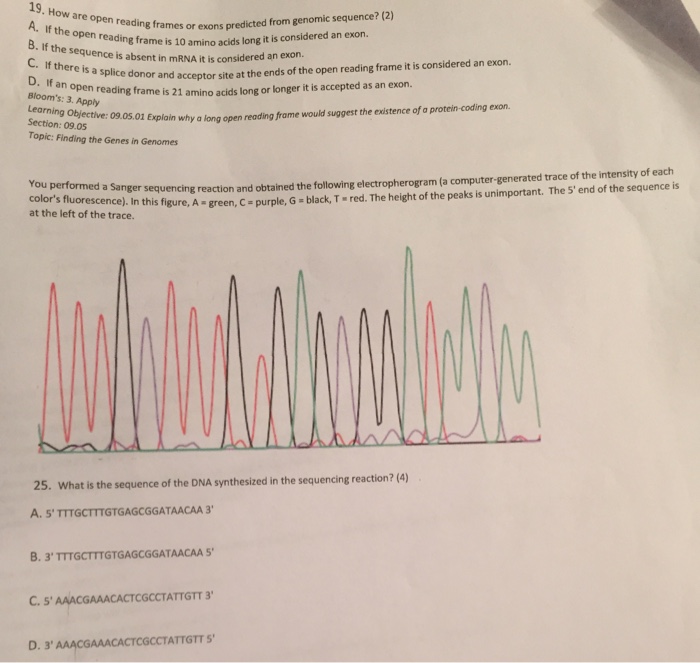
Show transcribed image text How are open reading frames or exons predicted from genomic sequence? A. If the open reading frame is 10 amino acids long it is consider an exon. B. If the sequence is absent in mRNA it is considered an exon. C. If there is a splice donor and acceptor site at the ends of the open accepted as an exon. D. If an open reading frame is 21 amino acids long or longer it is accepted as an exon. You performed a Sanger sequencing reaction and obtained the following electropheogram. In this figure, A = green, C = purple, G = black, T = red. The height of the peaks is unimportant. The 5' end of the sequence is at the left of the trace. 25. What is the sequence of the DNA synthesized in the sequencing reaction?
How are open reading frames or exons predicted from genomic sequence? A. If the open reading frame is 10 amino acids long it is consider an exon. B. If the sequence is absent in mRNA it is considered an exon. C. If there is a splice donor and acceptor site at the ends of the open accepted as an exon. D. If an open reading frame is 21 amino acids long or longer it is accepted as an exon. You performed a Sanger sequencing reaction and obtained the following electropheogram. In this figure, A = green, C = purple, G = black, T = red. The height of the peaks is unimportant. The 5' end of the sequence is at the left of the trace. 25. What is the sequence of the DNA synthesized in the sequencing reaction?



