Question: The following kinetic data were obtained from a protease that catalyzes cleavage of a substrate (…
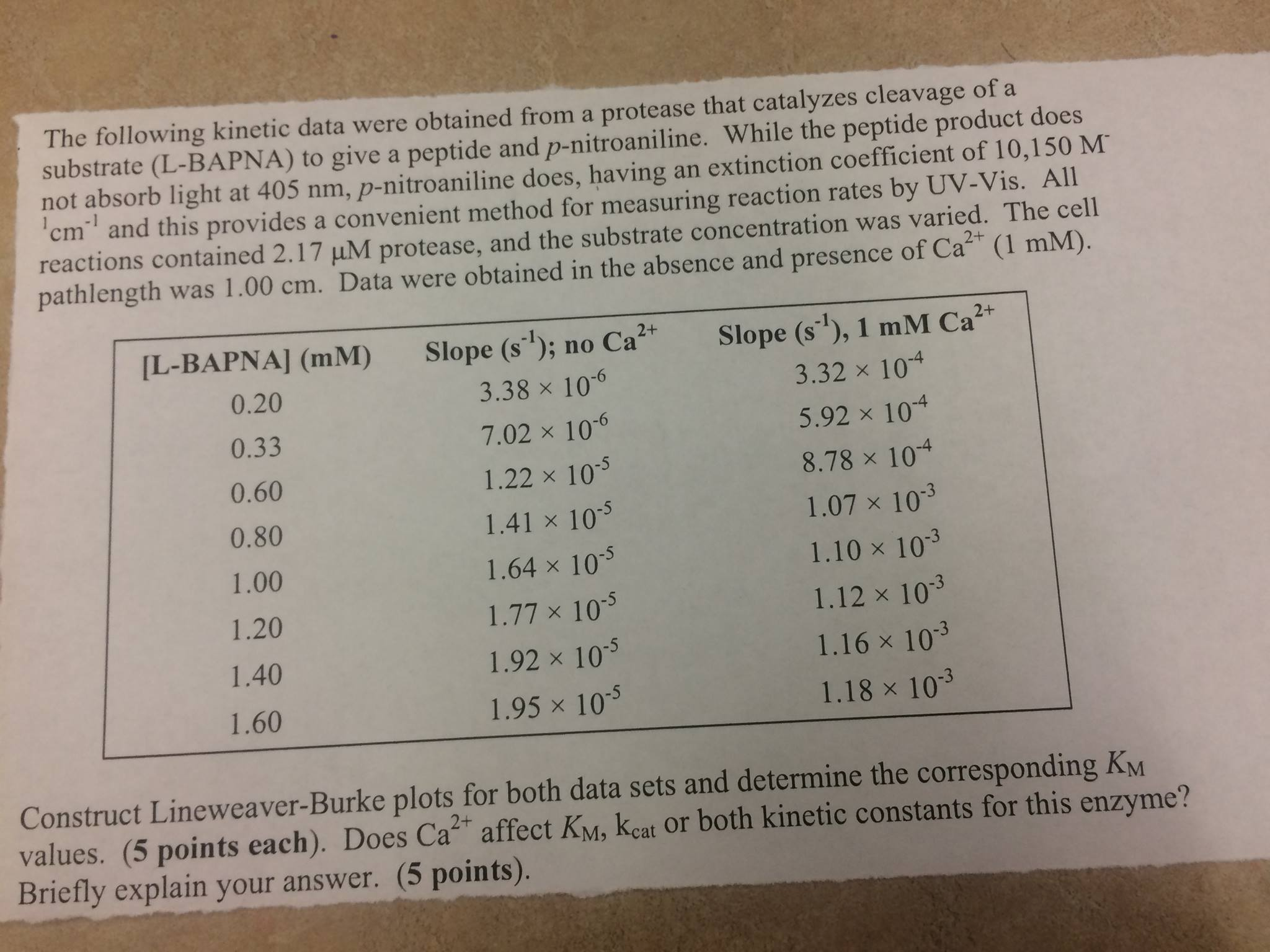
Show transcribed image text The following kinetic data were obtained from a protease that catalyzes cleavage of a substrate (L-BAPNA) to give a peptide and p-nitroaniline. While the peptide product does not absorb light at 405 nm, p-nitroaniline does, having an extinction coefficient of 10,150 M^- 1^cm^-1 and this provides a convenient method for measuring reaction rates by UV-Vis. All reactions contained 2.17 muM protease, and the substrate concentration was varied. The cell pathlength was 1.00 cm. Data were obtained in the absence and presence of Ca^2+ (1 mM). Construct Lineweaver-Burke plots for both data sets and determine the corresponding K_M values. Does Ca^2+ affect K_M, k_cat or both kinetic constants for this enzyme? Briefly explain your answer.
The following kinetic data were obtained from a protease that catalyzes cleavage of a substrate (L-BAPNA) to give a peptide and p-nitroaniline. While the peptide product does not absorb light at 405 nm, p-nitroaniline does, having an extinction coefficient of 10,150 M^- 1^cm^-1 and this provides a convenient method for measuring reaction rates by UV-Vis. All reactions contained 2.17 muM protease, and the substrate concentration was varied. The cell pathlength was 1.00 cm. Data were obtained in the absence and presence of Ca^2+ (1 mM). Construct Lineweaver-Burke plots for both data sets and determine the corresponding K_M values. Does Ca^2+ affect K_M, k_cat or both kinetic constants for this enzyme? Briefly explain your answer.



